Back to Home
Docker
Ligand Binding
Structure Prediction
FunFOLD5
Template-based ligand binding site prediction using structural alignment and quality assessment.
FunFOLD5
Powerful ligand binding site prediction using template-based structural alignment. Identify potential binding pockets with confidence.
Docker
Template-based
GitHub
🎯
Precision Binding Sites
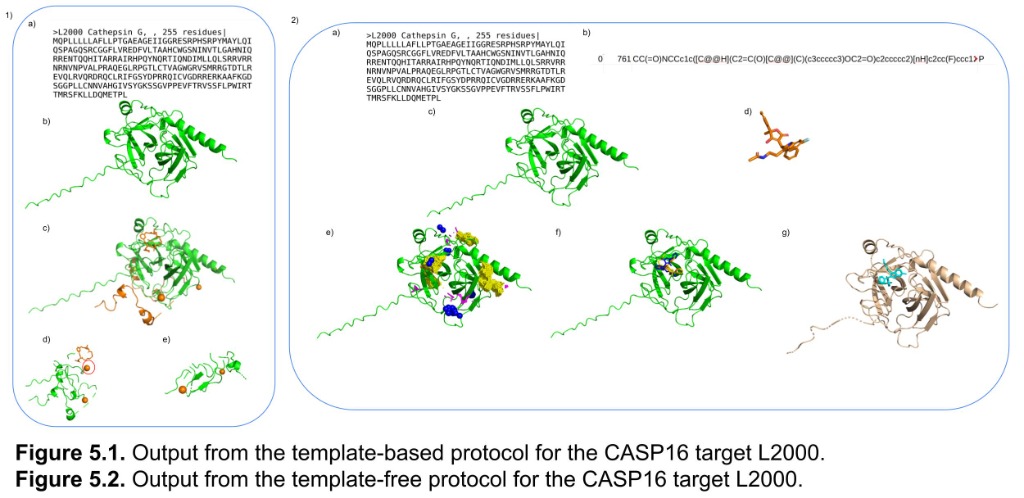
Template-based vs template-free protocols for CASP16 target L2000
🐳 Docker Installation
$ docker pull radiyaman/funfold5_template
🔬
Template-based
Leverages known protein-ligand structures
📍
Binding Sites
Accurately predicts ligand binding pockets
⭐
Quality Scoring
Ranks predictions by confidence
Usage Example
# Run FunFOLD5 on your protein
docker run -v $(pwd):/data radiyaman/funfold5_template \
-i /data/target.pdb \
-o /data/results
Methodology
1
Structural Alignment
Align target against template libraries
2
Binding Site Transfer
Transfer ligand binding information from templates
3
Confidence Scoring
Rank predicted sites by reliability